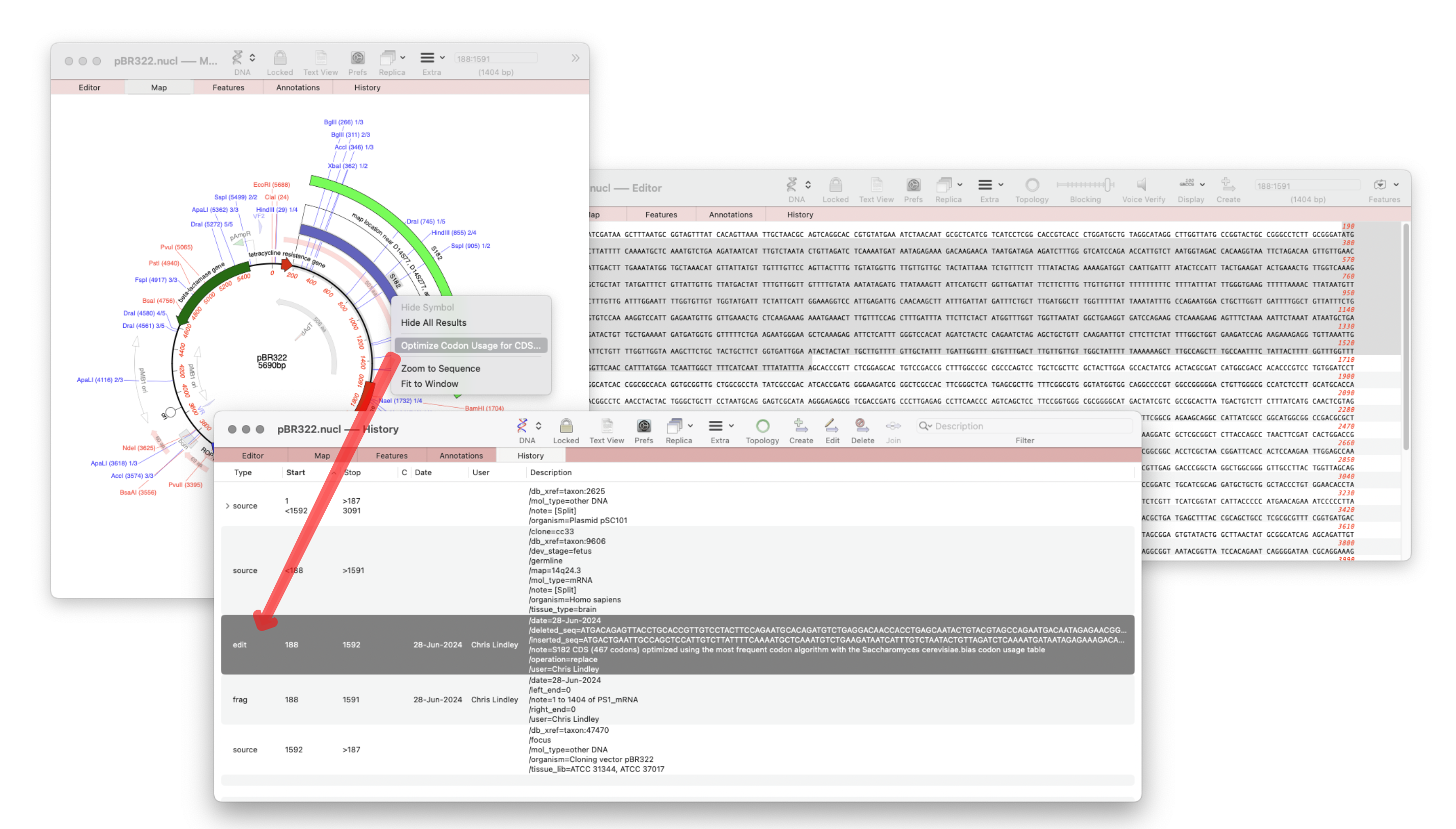
The recently released MacVector 18.7 has a new History tab in the Single sequence editor that shows the editing history of your DNA sequences
Since the introduction of MacVector’s Cloning Clipboard, all cloning actions (such as ligating a digested fragment into a vector) create a /FRAG feature that records the source of the ligated fragment, the restriction enzymes used to digest it (and any end treatment such as Klenow) as well as a timestamp. Then with the introduction of the OPTIMIZE CDS (MacVector 18.6) feature this also creates a new EDIT feature that details information about the action.
The new History tab lists these “history” features along with a timestamp and user of when they were performed thus allowing you to track the edit history of your sequences.
Although currently there are only a few edit operations that store information in the History tab, eventually all sequence modifications will write out this information allowing the full history of any construct to be determined and eventually allowing a simple roll back of the construct to how it existed on a specific date. The history tab may be sorted according to the user and date as well as the usual sort columns.

The following edit operations are currently shown in the History tab.
- CDS Optimization: creates an /EDIT feature.
- Cloning operations: such as ligating genes into a vector, create a /FRAG feature.
- SOURCE annotations: These are a mandatory Genbank feature that summarizes the length of the sequence, scientific name of the source organism, and Taxon ID number. Can also include other information such as map location, strain, clone, tissue type, etc., if provided by submitter. Note: Source features are a standard Genbank feature and do not include timestamp or user.
Unlike all other annotation/features EDIT and FRAG features are not standard Genbank nomenclature. However, if you need to export your sequence as “pure” Genbank you can use FILE | EXPORT | Genbank Flat (strict). This will remove all non standard terms and export a file that is 100% compliant with the Genbank specification.
How to view the EDIT history of your sequence.
- Perform a ligation of a digested fragment from another sequence
- Perform a codon optimisation of a CDS feature
- Switch to the HISTORY tab of your sequence.
MacVector 18.7 was released in July 2024 and introduces a History tab to track the construction of your expression vectors and clones. It also includes direct support for Codon Usage Tables, creating custom Codon Usage Tables and batch translation of CDS features. Additionally, MacVector 18.7 enhances Assembler’s toolkit by adding a new reference assembler for mapping PacBio and ONT sequencing reads to your reference sequences.