Here’s what MacVector can do for your lab.
|
Whatever type of alignment your sequence needs, there’s a tool in MacVector. 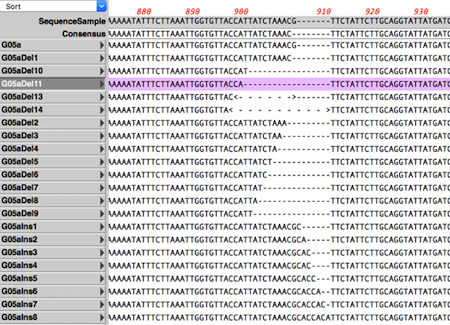
CRISPR Indel Analysis: Identify insertions and deletions following CRISPR editing of a target. Compare Genomes: Compares two related annotated genomes to identify identical, similar and weakly similar features. Sequence assembly of NGS data against a reference genome or compare your sequencing against your new construct. Coverage Tab: Compare different datasets assembled against the same reference sequence with expression level comparison. Translated Multiple Sequence Alignments: Align DNA sequences based on their translations. 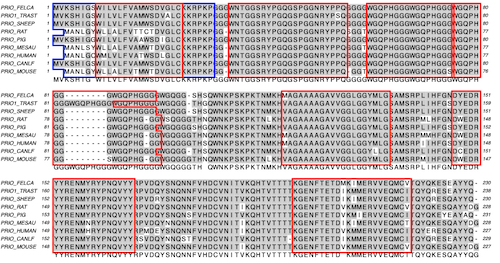
Visualise shared aligned domains: Protein alignments display domains that are conserved between sequences. Align proteins against a reference great for comparing known proteins against an unknown one. Auto Annotation of common plasmid features to blank sequences. InterProScan: Scan proteins for functional domains against many databases. 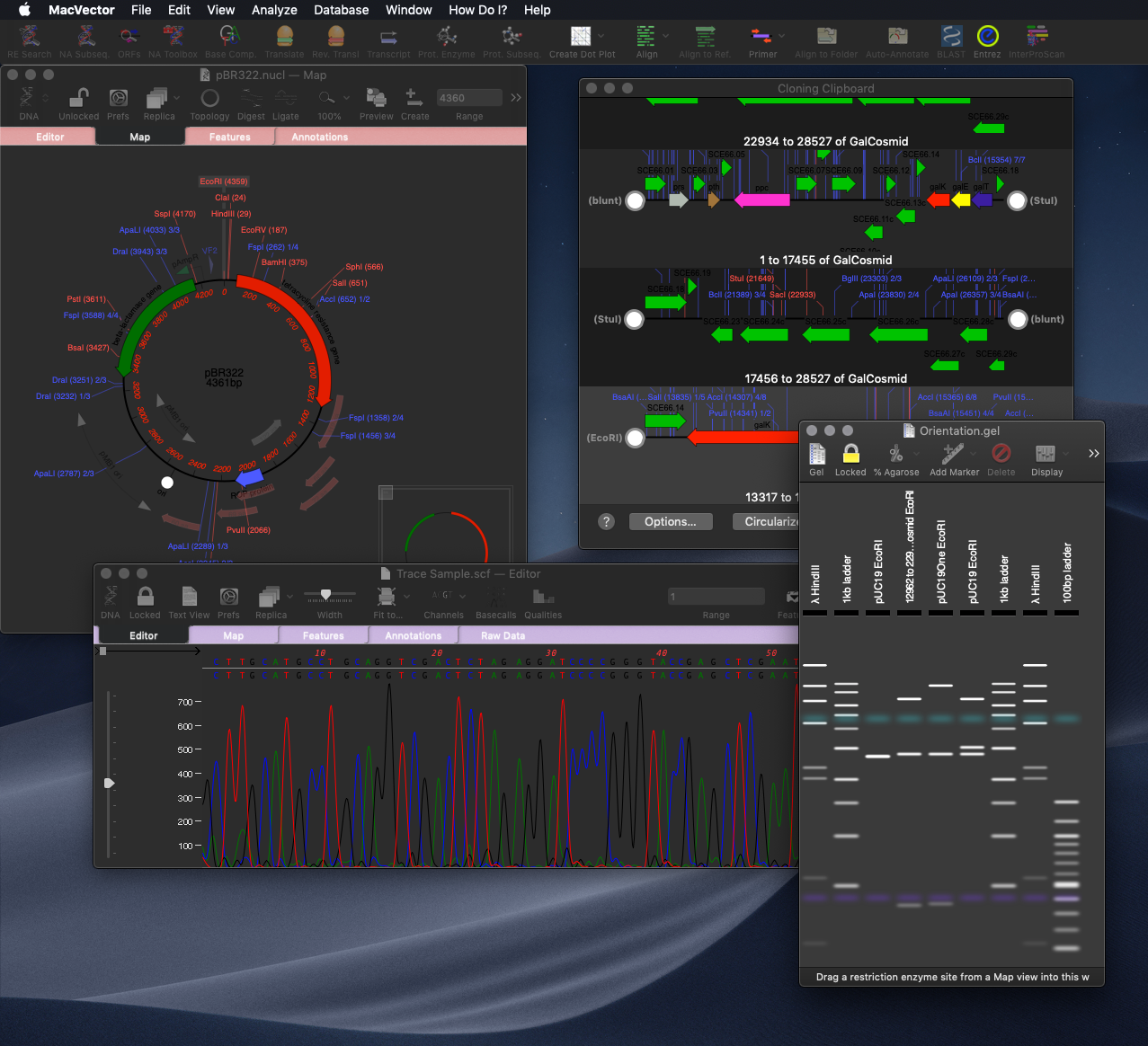
How Do I?: new menu shows common workflows with step by step guides. Every tool has a link to a video tutorial. |
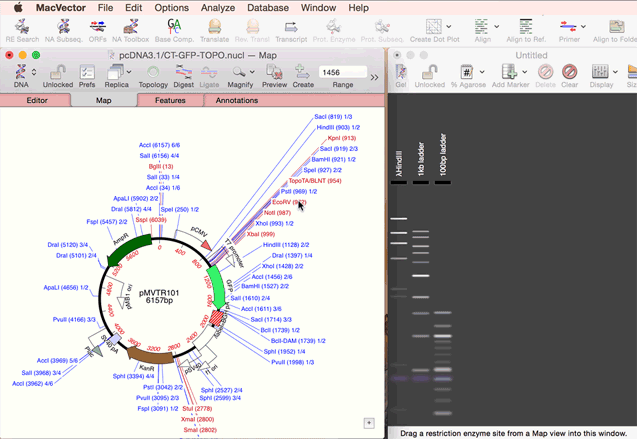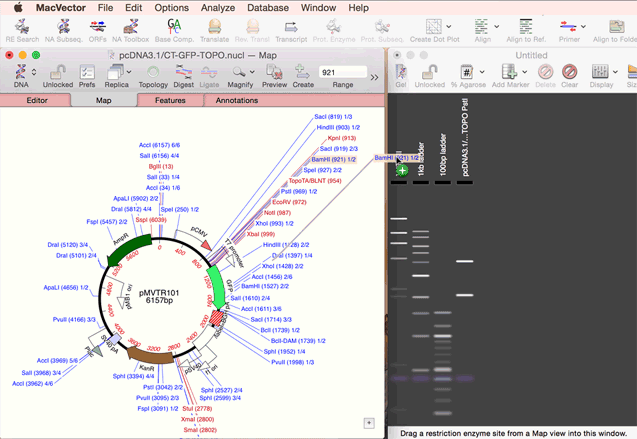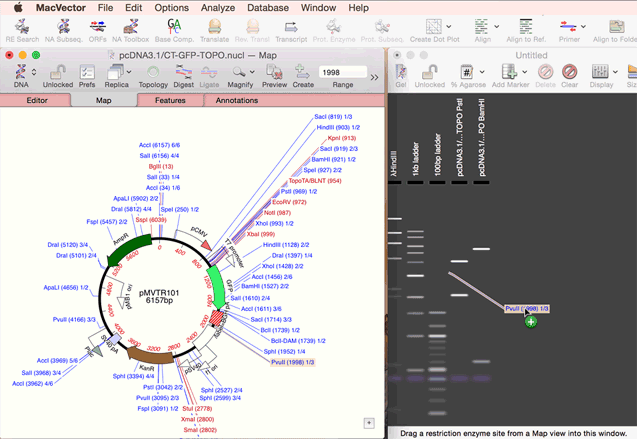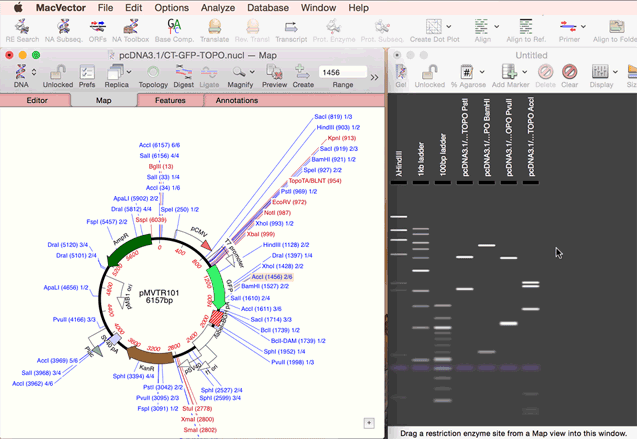
As simple as dragging a fragment to a cloning vector. Flexible Cloning Subclone with restriction enzymes, Gibson cloning, Gateway and more. Cloning history Every step is documented. Agarose Gel: run out digested sequences. Easily identify site(s) to differentiate successful clones. Restriction Enzyme Picker: easy identification of useful enzyme cut sites. 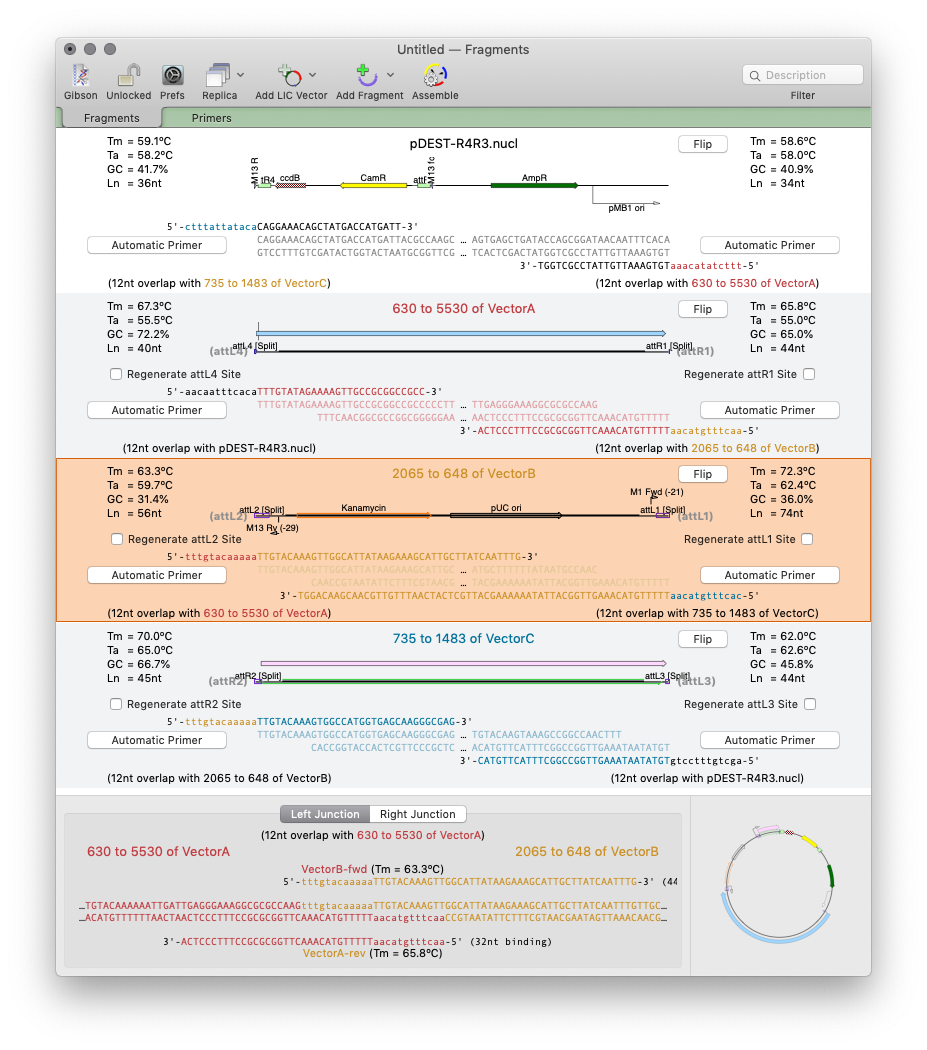
Gibson/Ligase-Independent Assemblies: Automatic primer design and assembly for Gibson Assembly or Ligase Independent Cloning workflows. Design primers with ease. QuickTest Primer changes primer design. Hairpin? Nudge your primer until it goes. Add tails to your primers with silent restriction sites/mismatches and view reading frame changes. Quickly design pairs of primers click a region to get the best primer pairs to amplify it. Scan For… Missing Primers: utilize the power of the Primer Database to display binding sites from your lab’s primer collection |
