Category: Techniques
-
Restriction enzyme analysis in MacVector and REBASE
Although there are two different ways to perform restriction enzyme analysis with MacVector, there are also additional places where restriction enzyme sites are shown. All these tools use the same set of restriction enzyme files to recognise enzymes. These files are updated regularly from the REBASE database. The restriction enzymes are divided into multiple files.…
-
Know Your Alignments
We often get asked “how do I do an alignment” using MacVector? Well, the answer to that is always “it depends”, and it depends on what you want to learn about your sequence(s). Here’s a quick summary of the different types of alignments and what you would use them for: You can learn more about…
-
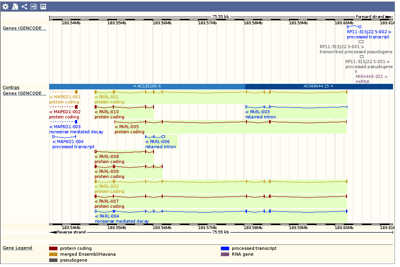
Importing sequences/features from websites such as ENSEMBL or UCSC’s Genome Browser
Many Genome Browsers/databases allow you to browse and view genomes, or a specific gene/region, with a high degree of detail. For example ENSEMBL and the UCSC Genome Browser. However, many times you want to work with that data on your own Mac. As usual the easiest way to exchange sequence data is using the Genbank…
-
A few tips on working with digested fragments in the Cloning Clipboard
The Cloning Clipboard is an easy, and flexible, way to design and document your cloning strategies. Here’s two tips on manipulating a single fragment.
-
Downloading BLAST Hits from the NCBI direct to your desktop
When you use MacVector to run a BLAST search, did you know that you can download any matching sequences directly from the BLAST Description List window? Simply select text on any part of the line(s) representing the sequence you want to download and choose Database | Retrieve To Desktop or Database | Retrieve To Disk…
-
Create constructs using the Cloning Clipboard
You can create new constructs in MacVector by selecting two restriction enzyme sites, choosing Edit -> Copy, selecting a target restriction site in a different molecule and then choosing Edit -> Paste. It works great and fully understands compatible overhanging sticky ends preventing you from accidentally creating biologically impossible molecules. However, a far more flexible…
-
Using the Primer Database to store your lab’s collection of primers
MacVector 14 saw the introduction of the new Primer Database tool. This allows you to save and retrieve primers from the Primer Database from the Primer3 and Quicktest Primer tools. You can also scan sequences for potential primer binding sites. The tool comes with a starter database of primers, but you can use existing subsequence…
-
Opening matching sequences from an Align To Folder search
You can use the Database->Align To Folder menu function in MacVector to quickly search any set of folders on your hard drive for matching sequences. Its like having your own personal BLAST search that just scans files of interest to you. The great thing is, when you find matching sequences, you can easily open them…
-
Quickly checking a small sequencing project
For analyzing large sequencing datasets you need Assembler. However, many times you do not need a powerful tool but just a quick way to check some sequencing data. For example for checking small sequencing projects, such as a site directed mutagenesis, looking for SNPs in a PCR product, cloning a gene or checking your latest…
-
Testing pairs of PCR primer
Over the past few releases of MacVector, we have changed primer testing considerably. Now all primer design and testing can be done using Quicktest Primer for a single primer and Primer Design (Primer3) for pairs of primers. If you want to test an existing pair of primers for suitability, use the following steps: A window…
