Tag: restriction digests
-
A few tips on working with digested fragments in the Cloning Clipboard
The Cloning Clipboard is an easy, and flexible, way to design and document your cloning strategies. Here’s two tips on manipulating a single fragment.
-
Create constructs using the Cloning Clipboard
You can create new constructs in MacVector by selecting two restriction enzyme sites, choosing Edit -> Copy, selecting a target restriction site in a different molecule and then choosing Edit -> Paste. It works great and fully understands compatible overhanging sticky ends preventing you from accidentally creating biologically impossible molecules. However, a far more flexible…
-
Removing a restriction site from a plasmid with the Cloning Clipboard
Being able to remove a restriction site by digesting with an enzyme that cuts at one site to linearize a plasmid, Klenow treating the linear fragment, then religating the fragment, is a useful trick in the lab. It can help you produce a suitable, and simple, cloning site when none previously existed in your vector.…
-
Keep up to date with MacVector auxiliary files
MacVector has a large collection of data files that are installed in folders alongside the MacVector application. These include Restriction Enzymes files, Scoring Matrices, documentation, tutorials, a variety of common cloning vectors and even a few useful Applescripts. If you use MacVector’s built-in automatic updater to install each new release and bug-fix, you will find…
-
How to select all identical restriction enzyme sites
The Automatic Restriction Enzyme Analysis tool that is displayed on the sequence in the Map tab is very powerful. It automatically displays a custom set of restriction enzyme recognition sites on every sequence that you open. Unique recognition sites are displayed in red and sites with two or more cut locations are displayed in blue.…
-
Restriction enzyme sites in cloning vectors show compatible ends with fragments in the Cloning Clipboard
If you are looking for compatible cloning sites for a fragment, MacVector has a simple color-coded function to identify potential target sites in a vector. First select the fragment you wish to clone by clicking on two enzymes in the Map view of a single sequence document or from a restriction enzyme search result. Then…
-
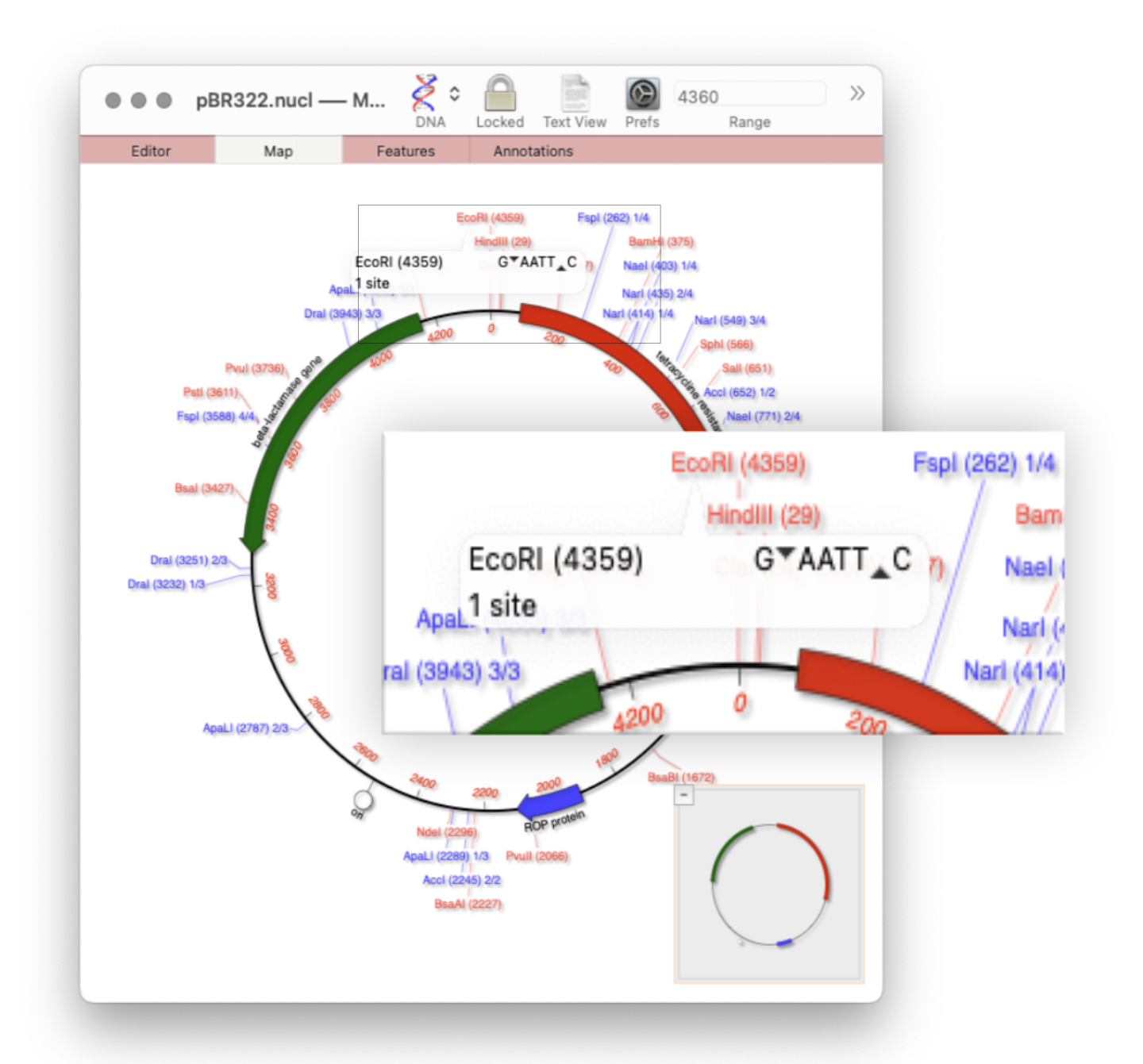
Restriction enzyme sites and tooltips
Quickly viewing the recognition sequence and cut site of a restriction site is very easy in the Map tab. If you hover your mouse over a restriction site in the Map tab, a tooltip will show you the restriction enzyme recognition site, the location of the cut site, and number of times that enzyme cuts…
-
Use the [option] key to paste, or ligate, in the opposite orientation
The most simple way of generating new constructs in MacVector using restriction enzyme sites is to: MacVector is intelligent enough to understand compatible sticky ends during this procedure and will prompt if they do not match. For a simple cloning (e.g. inserting a copied EcoRI fragment into the EcoRI site of a vector), by default,…
-
Simulated Agarose Gels
MacVector 14.5 has a Agarose Gel interface which allows you to view photo-realistic recreations of restriction digests of linear and circular DNA molecules. The gels look so realistic that users have had a hard time telling photos of their own digests from the simulation in MacVector. When you first use the new tool and compare…
-
MacVector Cloning Edition
Does your lab only need a simple DNA analysis application that will help you design primers and cloning experiments? Has your budget been cut to the bone? If the answer is a resounding “yes”, then we’ve released a new version of MacVector just for you. MacVector Cloning Edition provides all of the functionality you need…
