Gibson/LIC Assembly
MacVector has a unique interface that lets you design and document cloning experiments using the popular Gibson Assembly and Ligase Independent Cloning (LIC) techniques that do not require the use of Restriction Enzymes or T4 DNA Ligase. Both use similar approaches, where you design primers to amplify two or more fragments with 15-20nt overlaps at their ends that can subsequently be assembled into circular molecules with the help of treatment with 5' exonuclease (Gibson) or the 3' exonuclease activity of T4 DNA Polymerase under controlled conditions (LIC).
Overview
MacVector uses a highly interactive project interface as a base for this functionality. You create a project, telling MacVector what sort of technique you intend to use, then add the fragments of DNA you would like to join together and MacVector will automaticallygenerate a suitable list of balanced primers for you. You can ask for extra spacer residues, to adjust open reading frames for example, or supply your own custom primers that can have mismatches if required. Once you are happy with the design, you can assemble the fragments into a circular product, complete with all of the original annotations, and print/save/copy the list of required primers. Any individual PCR fragment can be saved or copied, again complete with annotations. Finally, you can also supply MacVector with a collection of pre-designed PCR fragments with overlapping ends and MacVector will assemble those into a circular product.
Gibson Assembly
To start a new project, select File | New | Gibson/Ligase-independent Assembly..., then choose the first option as shown below and click on the Create button.

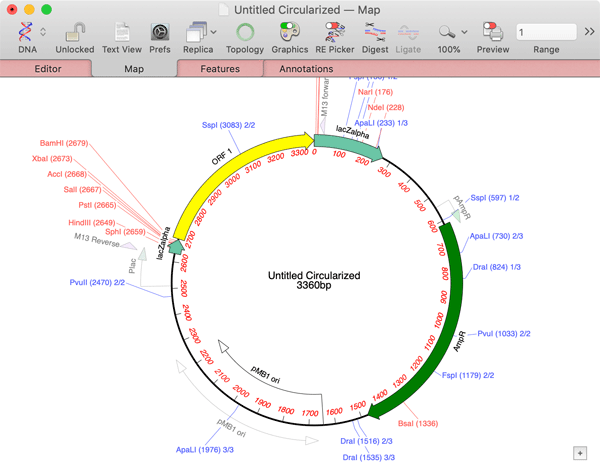
A new empty project window appears. There are many ways to add fragments to the window. They can be copy/pasted from other MacVector windows, or opened from files on disk. One of the easiest ways to add fragments is via drag and drop. For example, one common practical approach for Gibson Assembly is to use a simple vector cut with a restriction enzyme as the core vector sequence, avoiding the need for an additional PCR reaction. You can use this approach in MacVector by opening the vector of interest (pUC19 in this case), selecting the restriction enzyme you want and dragging that site into the Gibson project window. This example uses the enzyme SmaI that generates blunt ended fragments;
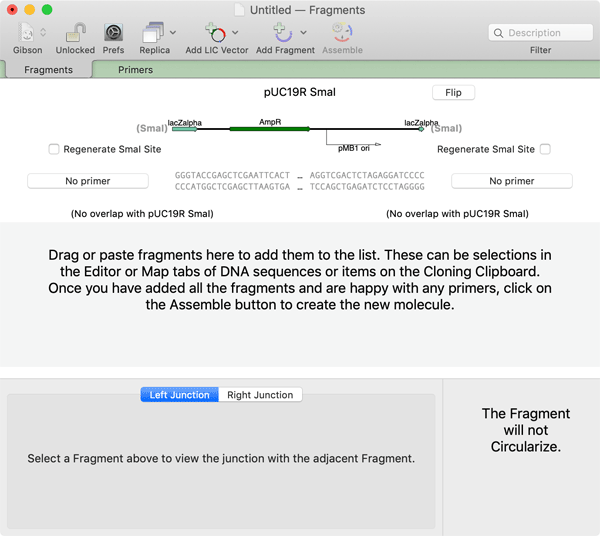
Now you can add required fragments to the Assembly. In this example, we just selected a CDS feature from the Map tab of a sample file and dragged that into the project;
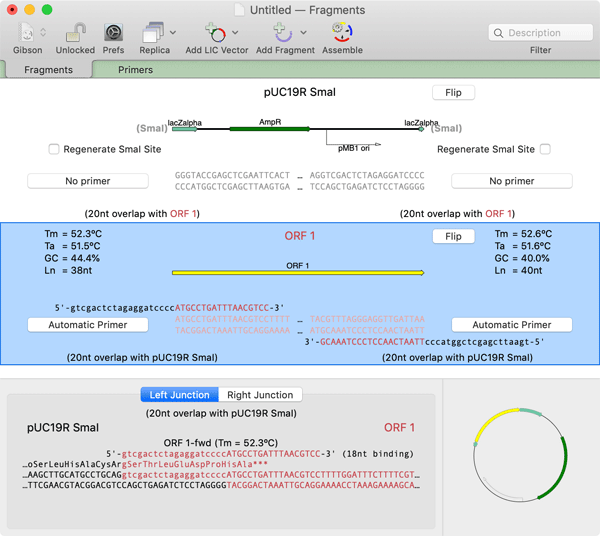
Once the second fragment is added, MacVector automatically calculates a suitable pair of primers. Because the vector is supplied as a restriction fragment, the primers have 20nt "tails" to provide the required 20nt overlap. The "core" of the primer, where it binds to the ORF 1 DNA for the PCR reaction, by default is a minimum of 18nt. In this case, the right hand primer actually has a core binding of 20nt in order to provide a balanced Tm for the PCR pair.
When a fragment is selected, there is a lower details pane that shows the left and right junction of the fragments with its adjacent fragments. This also shows the translations of CDS features that are present near the junctions. In this example, we can see that the CDS coming into the junction from the left hand side (the beta-galactosidase alpha gene CDS) is not actually in-frame with the ATG codon from ORF 1;
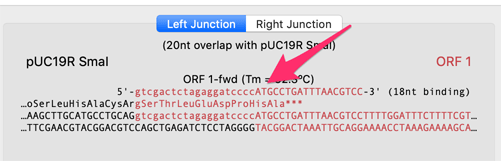
We can ask MacVector to insert additional residues into the primer so that the two CDSs become fused in-frame. if you click on the Automatic Primer button at the left end of the ORF 1 fragment, an editing pane appears;
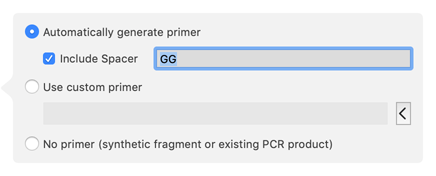
The Left Junction display updates in real time as we type in the Include Spacer box, so we can see that adding two additional residues is required to get the two CDS features in-frame;

The Primers tab displays a list of the required primers that can be printed, saved as an Excel compatible file or added to the built-in MacVector Primer Database;

At any time, the entire project can be saved to disk. In addition, a context-sensitive menu item lets you save left and right primers, and also any individual fragment as its predicted PCR product, complete with tails and any mismatches, so that you have a record of the exact sequence of each of the components of your construct.
Finally, you can click on the Assemble toolbar button to create a fully annotated product.
Ligase Independent Assembly
While there are a number of variants of this, one of the most popular approaches takes advantage of the 3' exonuclease activity of T4 DNA Polymerase to create long 5' overhangs at the ends of fragments. The key here is that if the ends are missing a specific dNTP for ~15nt or more, you can treat the linearized molecule with T4 DNA Polymerase in the presence of the missing dNTP, and the ends will be cut back to the first occurence of the dNTP, potentially leaving long 5' single stranded overhangs. This greatly facilitates the assembly process so that much shorter overlaps are sufficient for high efficiency circularization.
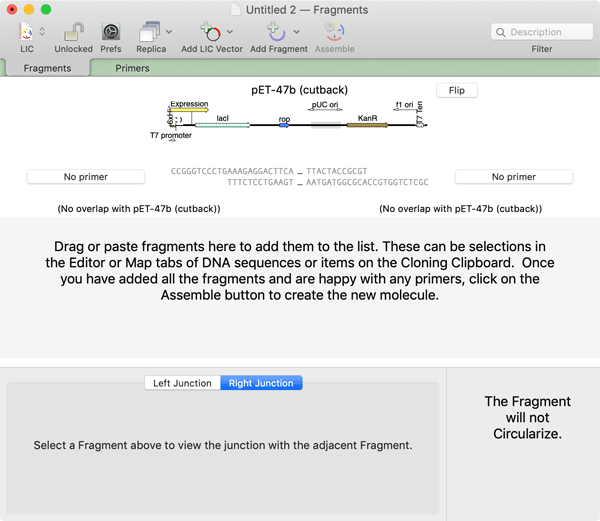
A variety of vectors have been identified that fortuitously happen to have suitable cloning sites, while others have been specifically engineered. MacVector includes a selection of LIC vectors in the /Applications/MacVector/Common Vectors/LIC Vectors/ folder. A convenient Add LIC Vector button on the toolbar lets you select one of these, or you can add your own from a file;
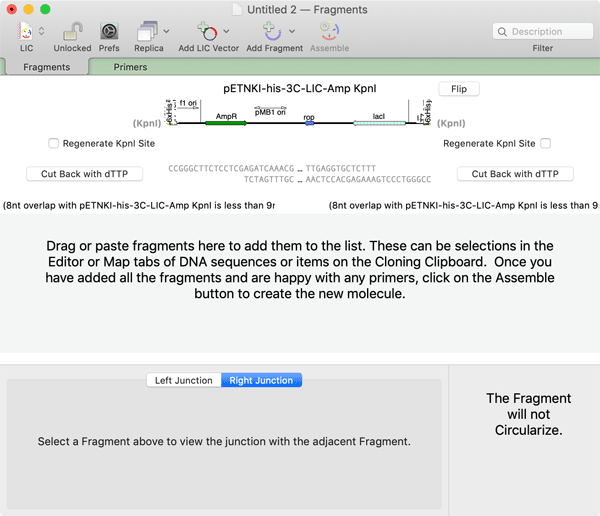
Most of the vectors already have cut-back ends, like the one above. But you can also generate the cutback ends yourself from any linearized vector, as with this KpnI-linearized vector cut back in the presence of dTTP;
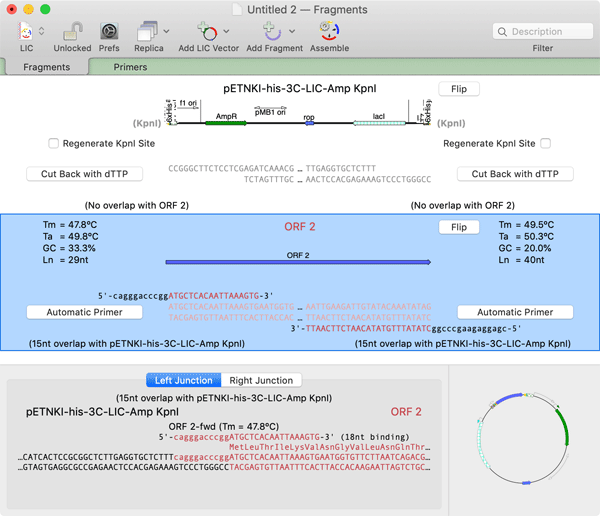
Again, you can add fragments and suitable primers will be generated. In this case, after PCR, the fragment would be treated with T4 DNA Polymerase in the presence of dATP in order to generate compatible single-stranded complementary ends.
Assembling Existing Fragments
You can also assemble PCR fragments that you have already created, either within MacVector or through other online interfaces such as NEBuilder;

|

2x.png)


2x.png)
