New Features in MacVector 12.0
Overview
MacVector 12.0 has a large number of major changes to better support circular sequences, enhance the sequence editor and improve the graphical display of features, in addition to a large number of minor enhancements. You may want to read the blog article discussing the new release.
Circular Sequence Support
While MacVector has always been able to display circular sequences, and some of the algorithms (e.g. Restriction Enzyme analysis) correctly reported results crossing the circular origin, previous versions of MacVector had limitations handling circular sequences. In particular, selections and features could not cross the circular split point. This has been resolved for MacVector 12 and most algorithms now seamlessly handle circular sequences and selections that cross the origin.
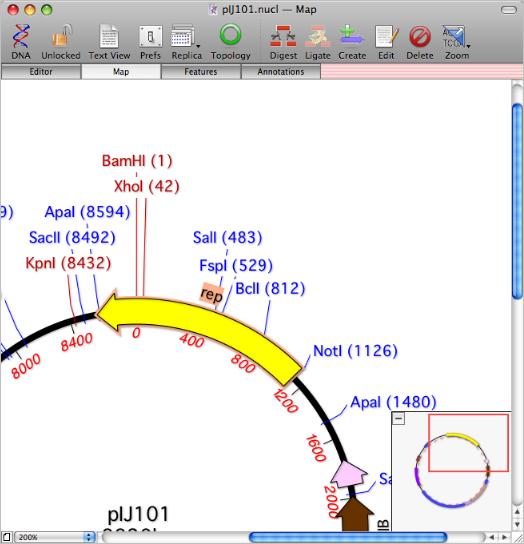
Screenshot showing selection of a feature crossing the circular origin, unique restriction enzymes displayed in red and the new navigation overview pane in MacVector 12.0.
Feature Map Enhancements
There have been a host of improvements to the Feature Map designed to improve navigation so you can quickly zoom in and out to view the detail of long sequences, along with a much improved circular display to match the general improvement in circular sequence support. A new overview pane lets you see exactly where you are at in larger sequences and the floating Graphics Palette window has been redisgned, adding a variety of new buttons to aid navigation. There are also some new graphical types that can be used to display features and the sequence itself can now be displayed double-stranded. You can also now display the translations or sequence of certain feature types (e.g. CDS or primers) underneath or on top of the sequence when zoomed to the residue level.
Restriction Enzyme Enhancements
Enzymes that cut a sequence just once are now displayed in red so you can quickly identify unique cutters in the Map view. When zoomed to the residue level, the staggered ends of any cuts sites are clearly displayed on the sequence and are highlighted appropriately when a Restriction Enzyme fragment is selected.

You can now see double stranded sequence in the Map view, including the staggered ends produced by restriction enzyme selections, plus the translations of CDS features can be viewed with the DNA.
If you have a Restriction Enzyme fragment copied to the clipboard, any enzymes with compatible sticky ends are shown with a colored background indicating which end of the clipboard fragment they are compatible with.
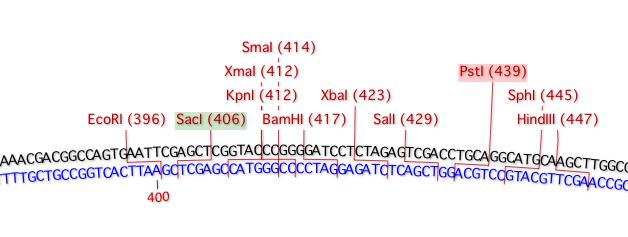
With a SacI - PstI fragment on the clipboard, compatible sites in a target molecule are shown with colored backgrounds to simplify the selection of suitable ends for ligating the fragment.
You can now search for "one out" sites that might be useful targets for site-specific mutagenesis experiments or for use in primer design.
Sequence Editor Changes
By popular demand, you can now have a mix of upper and lower case residues in the main sequence editor. In addition, you can now color the residues or background across regions of interest in the Editor. This is tightly linked to the Features in the Map view so you can quickly and easily create colored segments to be displayed in both views.
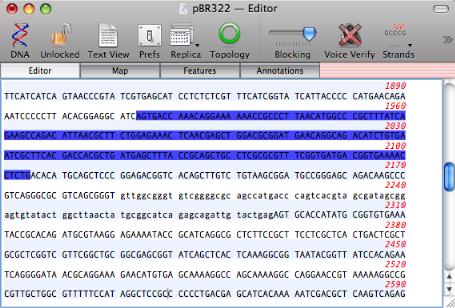
Miscellaneous Changes
As usual with our releases, there are a large number of minor enhancments designed to streamline your everyday use of MacVector. A full list of changes in included in the Release Notes.
|

2x.png)


2x.png)
