Tag: cloning
-
How to select all identical restriction enzyme sites
The Automatic Restriction Enzyme Analysis tool that is displayed on the sequence in the Map tab is very powerful. It automatically displays a custom set of restriction enzyme recognition sites on every sequence that you open. Unique recognition sites are displayed in red and sites with two or more cut locations are displayed in blue.…
-
Restriction enzyme sites in cloning vectors show compatible ends with fragments in the Cloning Clipboard
If you are looking for compatible cloning sites for a fragment, MacVector has a simple color-coded function to identify potential target sites in a vector. First select the fragment you wish to clone by clicking on two enzymes in the Map view of a single sequence document or from a restriction enzyme search result. Then…
-
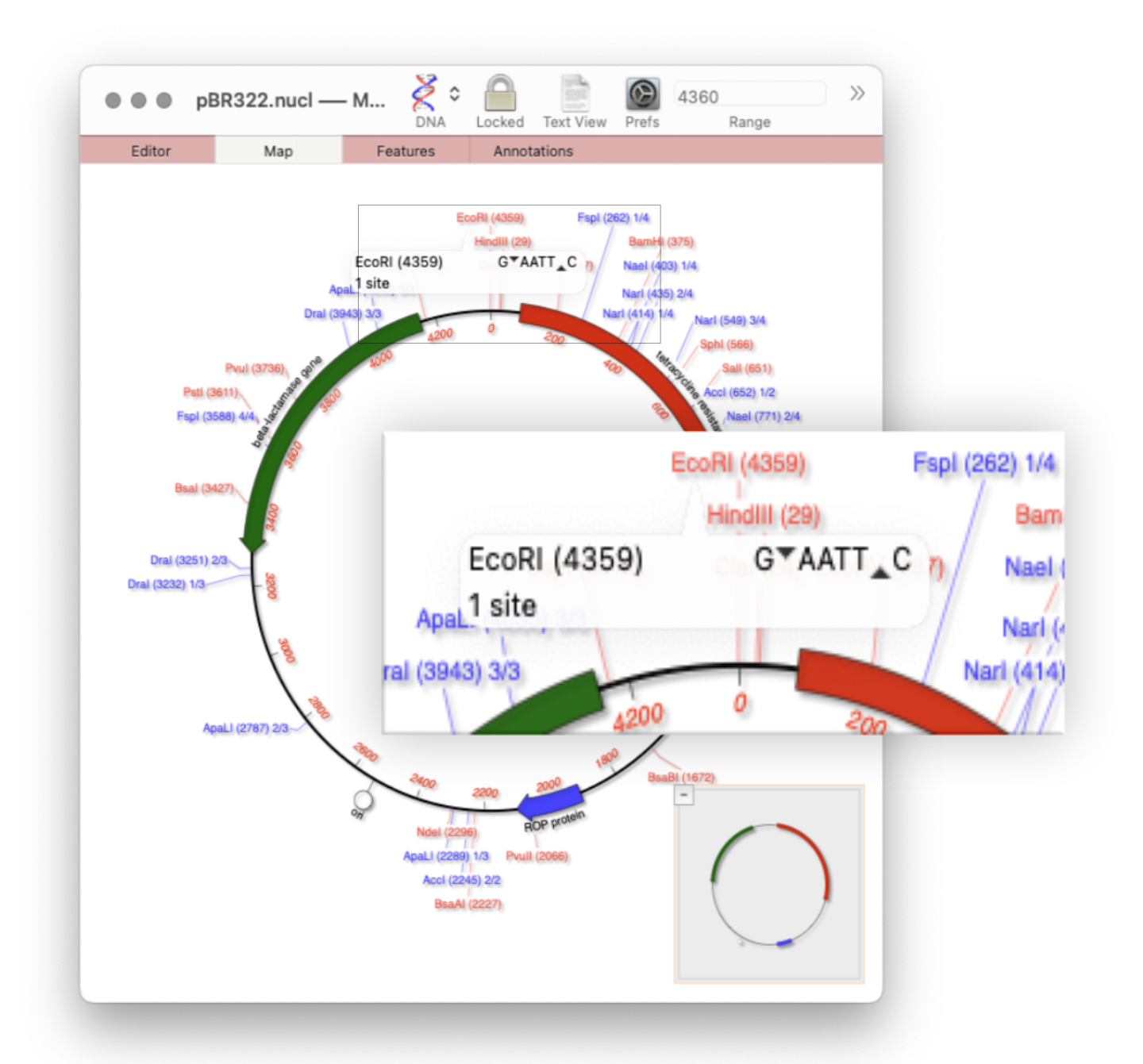
Restriction enzyme sites and tooltips
Quickly viewing the recognition sequence and cut site of a restriction site is very easy in the Map tab. If you hover your mouse over a restriction site in the Map tab, a tooltip will show you the restriction enzyme recognition site, the location of the cut site, and number of times that enzyme cuts…
-
MacVector for Windows: update.
It’s quite a few months since the last update on the development of MacVector for Windows. Work is progressing very well and a lot of the underlying functionality is now finished. The Cloning Clipboard, Restriction Enzyme analysis and QuickTest Primer tools are all done. The release is still some time away, but if you are…
-
Use the [option] key to paste, or ligate, in the opposite orientation
The most simple way of generating new constructs in MacVector using restriction enzyme sites is to: MacVector is intelligent enough to understand compatible sticky ends during this procedure and will prompt if they do not match. For a simple cloning (e.g. inserting a copied EcoRI fragment into the EcoRI site of a vector), by default,…
-
Creating cloning construction flowcharts in third party applications
We’ve previously discussed how every ligation is documented. You get a “Frag” annotation that contains the date, source sequence, enzymes used and any end modification that was done to that fragment during the ligation. However, we had regular requests to make it easier for user to document their constructs in other ways. For example being…
-
Simulated Agarose Gels
MacVector 14.5 has a Agarose Gel interface which allows you to view photo-realistic recreations of restriction digests of linear and circular DNA molecules. The gels look so realistic that users have had a hard time telling photos of their own digests from the simulation in MacVector. When you first use the new tool and compare…
-
Cloning Clipboard: A few tips on working with digested fragments
The Cloning Clipboard is an easy, and flexible, way to design and document your cloning strategies. Recently we discussed how every ligation in a cloning procedure is documented and stored in a sequence, so you always know how a construct was made. If you need to manipulate a fragment as a sequence, before ligating it,…
-
Cloning Clipboard: Documenting the history of a construct
Designing and documenting cloning strategies is easy with the Cloning Clipboard. You can perform quite complex ligations by simply dragging compatible ends together. Not only that but every step is documented and recorded in the resulting sequence, so you always know where each fragment came from. Ligating a single fragment into a vector is as…
-
MacVector Cloning Edition
Does your lab only need a simple DNA analysis application that will help you design primers and cloning experiments? Has your budget been cut to the bone? If the answer is a resounding “yes”, then we’ve released a new version of MacVector just for you. MacVector Cloning Edition provides all of the functionality you need…
