With the advent of cheap Next Generation Sequencing technologies, there has been an explosion of whole genome sequences deposited in BLAST databases. One consequence of this is that, particularly for sequences of bacterial origin, most of the significant hits are to entire genomes. The classic BLAST results show the sequence alignments, but give no indication of what features are present on the database sequence at the alignment location. MacVector has a new unique BLAST Map result tab that shows the actual annotations around the alignment location for each high scoring segment pair.
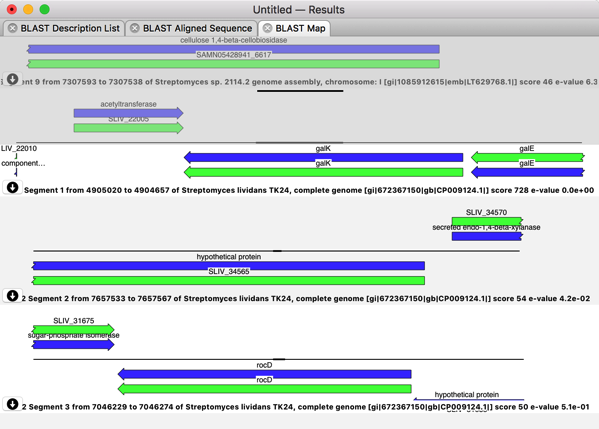
If a sequence is highly annotated, such that not all of the features are visible, simply holding the mouse over the appropriate pane will expand the pane to display all of the features. If you have already annotated (or partially annotated) your query sequence, those features will appear in the grey background “overlay” pane – that stays locked to the top of the window while the hits can be scrolled underneath it.