MacVector 18.7 has just been released. If you are eligible for this release you will be prompted to upgrade, otherwise go to MACVECTOR | CHECK FOR UPDATES… and follow the prompts to be automatically upgraded. If your license is not eligible then why not upgrade?
Overview
MacVector 18.7 introduces a History tab to track the construction of your expression vectors and clones. It also includes new features for codon optimization, such as direct support for Codon Usage Tables (CUT/.bias) and the ability to create custom Codon Usage Tables files from your own sequences. Additionally, MacVector 18.7 enhances Assembler’s toolkit by adding a new reference assembler for mapping PacBio and ONT sequencing reads to your reference sequences.
MacVector 18.7 also makes it easier to store your own Restriction Enzyme files when multiple people use the same Mac (ideal for that shared lab Mac!).
..and as usual there are a lot of enhancements to existing features such as protein pI calculations.
MacVector 18.7 was developed on macOS Sonoma and is supported on macOS High Sierra to macOS Sonoma. It has also been tested on early development releases of macOS Sequoia and will be fully supported when Apple release it. MacVector 18.7 is a Universal Binary that will run on Apple Silicon Macs and Intel Macs.
Long-Read Reference Alignments using minimap2
The addition of Minimap2 allows Assembler to map noisy long-read data from Pacific Biosciences or Oxford Nanopore to a reference sequence(s). Minimap2 is similar to Bowtie2 but optimized for handling long reads instead of short reads under 500 nucleotides. Additionally, Minimap2 also excels at assembling short read data and may even out-perform Bowtie2 in certain situations.
Translate All CDS Features
You can now easily translate all CDS features in a sequence with the new menu option Analyze | Translate All CDS Features. This is useful for translating proteins in bacterial genomes or eukaryotic sequences. You can choose to display all translated proteins in fasta format or create a codon usage table from the results.
Translate All CDS Features in Folder
There is a new Database | Translate All CDS Features in Folder menu option that is similar to Translate All CDS Features except that it takes a source folder and then loads every sequence file in the folder and translates each CDS feature that it finds, accumulating the results and offering the same result options as Translate All CDS Features. A Codon Usage Table viewer window is always created and displayed when you select this option.
Codon Usage Table Viewer
MacVector now includes a viewer for codon usage tables (CUT/.bias) files. This displays the data in a standard text format with one row of data per codon, identical to codon usage output windows used in other MacVector translation functions.

You can import Codon Usage Tables from CUT files available on codon usage websites such as CUTG. You can also generate custom CUT files by using the new Translate All CDS Features from a single sequence or from multiple sequences using the new Translate All CDS Features in Folder tool. You can invoke this multiple times and slowly build up the codon usage information from a large sequence data set in multiple folders.
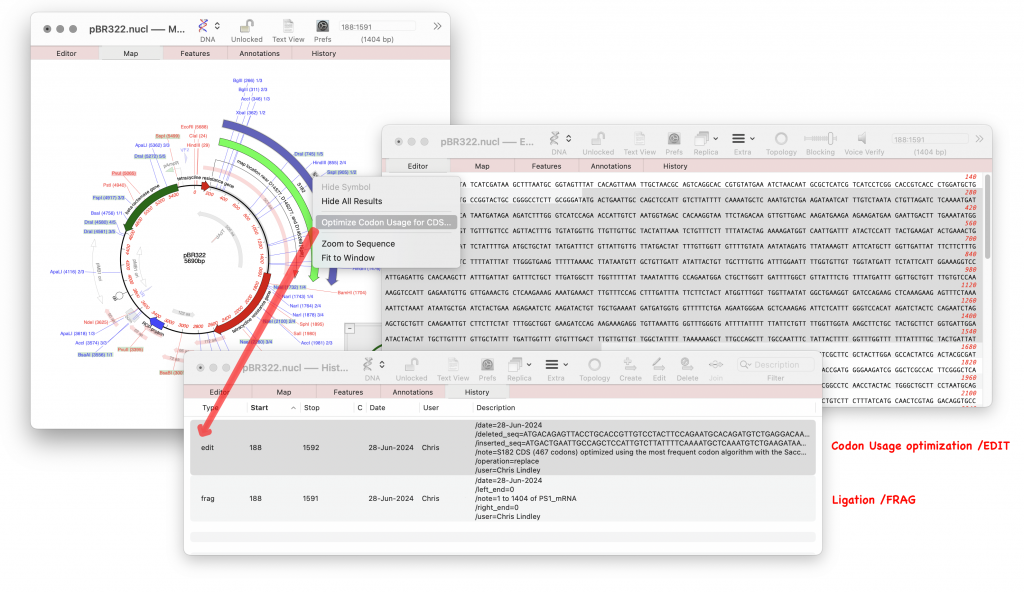
History Tab
There is a new tab in nucleic acid single sequence editors called History. This tab lists several MacVector-specific features relating to the editing history of the sequences such as ‘frag’ and ‘edit’. It also includes Source annotations which summarize the length of the sequence, scientific name of the source organism, and Taxon ID number. Apart from Source annotations these features now contain additional information such as the date of the operation, the name of the user who performed it and additional sequence information. In the future, all MacVector sequence modifications will write out this information allowing the full history of any construct to be determined and even allowing a simple reversion of the construct to how it existed on a specific date
Change in Default Restriction Enzyme File Location
MacVector now saves restriction enzyme files in a new location (~/Library/Application Support/MacVector/Restriction Enzymes/) within a user’s home folder. This is always writeable, even without Administrator access. If you have already saved files in a different location, they will not be affected. MacVector will automatically populate the new directory with the latest restriction enzymes and update any user-edited files from the old location.
Miscellaneous Enhancements and Bug Fixes
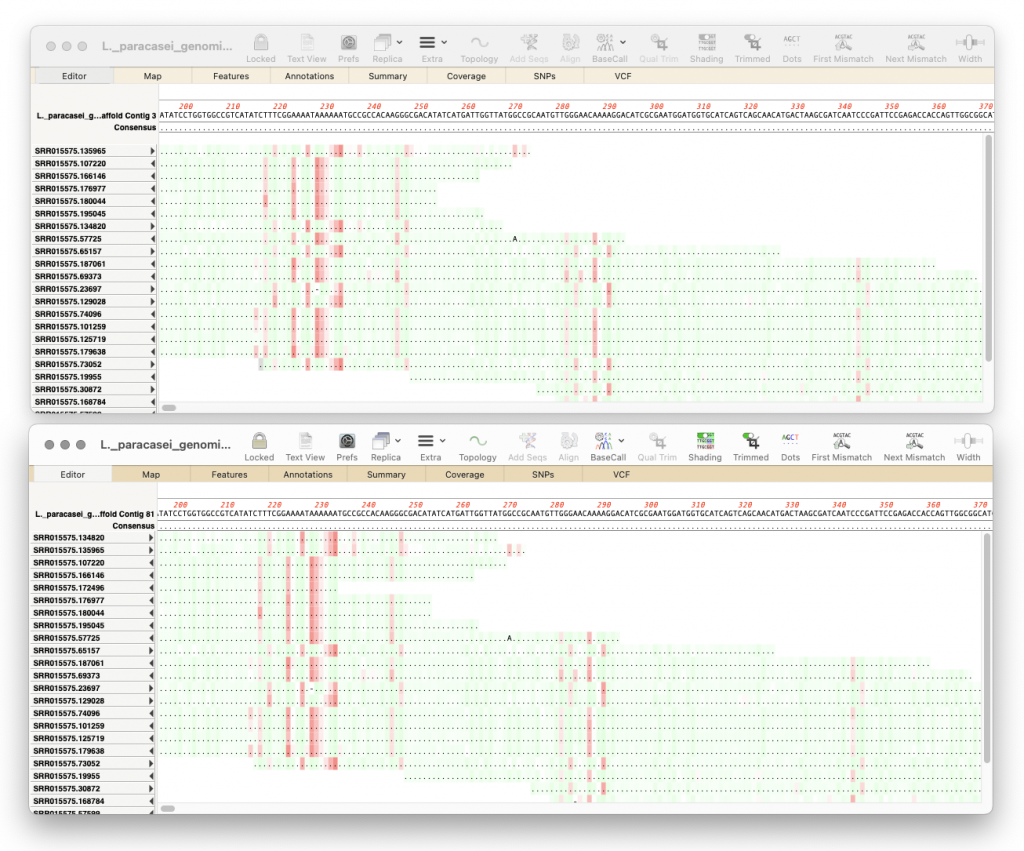
- The Align to Reference SNPs tab now also displays the percentage of each residue present in each heterozygote SNP.
- The Align to Reference consensus calling threshold default has been raised to 70% so that heterozygous SNPs are more consistently reported on the consensus line.
- A crash when repeating heterozygote analysis has been fixed.
- Copied fasta text data is now more reproducibly parsed as single sequence data by New From Clipboard.
- The protein pI calculations have been modified to also report the pI ignoring Trp and Cys residues. This brings the results more in agreement with the popular ExPASY website.
- A bug where the “blocking” for protein sequences was taking the DNA values has been fixed.
- Exporting sequence data in the Sequin .tbl format now correctly writes out the correct sequence for the minus strand of segmented features.