If the graphics in a nucleic acid sequence Map tab appear somewhat “washed out” it is because the graphic items represent common features that MacVector has found that are not annotated on the sequence. For example, here are the Map and Feature tabs of an unannotated cloning vector;
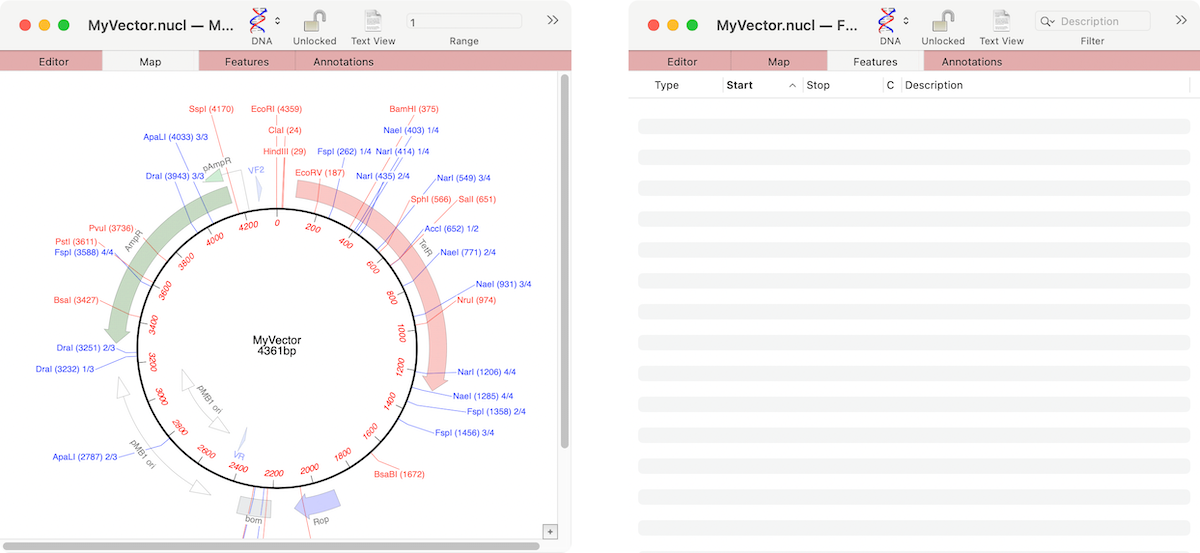
You can see a number of features on the Map tab, but the Features tab is completely empty. The graphics indicate common features that MacVector has identified that have not been annotated on the sequence. If you select one of the features in the Map tab and right-click (or [ctrl]-click) there is an option in the resulting context sensitive menu to Add CDS Feature. When that is selected, the feature takes on a bold appearance and a new annotation appears in the Features tab.

If you wish, you can select multiple missing features and then add them all with a right-click. Or you can select the Results | Missing Features tree view item in the floating Graphics Palette to select all missing features and then add with a right-click.
Note that the automatic display of missing features is controlled by the MacVector | Settings | Scan DNA tab. From there you can control how they are identified or even point the algorithm to your own folder of annotated sequences to be a source for the missing features.