Transposon mutagenesis is a common approach for investigating gene function in bacterial genomes by selecting for clones where the transposon inserting into the genome has generated a specific phenotype. You can then simply sequence the entire genome of each clone by NGS to identify the transposon insertion site. To lower the cost of such experiments, it is common to pool several individual genomes into each NGS sample and then run appropriate sequence analysis to identify the genes disrupted by the transposition events.
There is a new Transposon Insertion Analysis Tutorial that describes how to perform this analysis using MacVector with Assembler. To follow along, you can download sample data. The basic strategy is to use MacVector’s Align to Folder functionality to pull out all pairs of reads that contain transposon sequences then align those to the genome to identify the end points of the transposon insertion site.
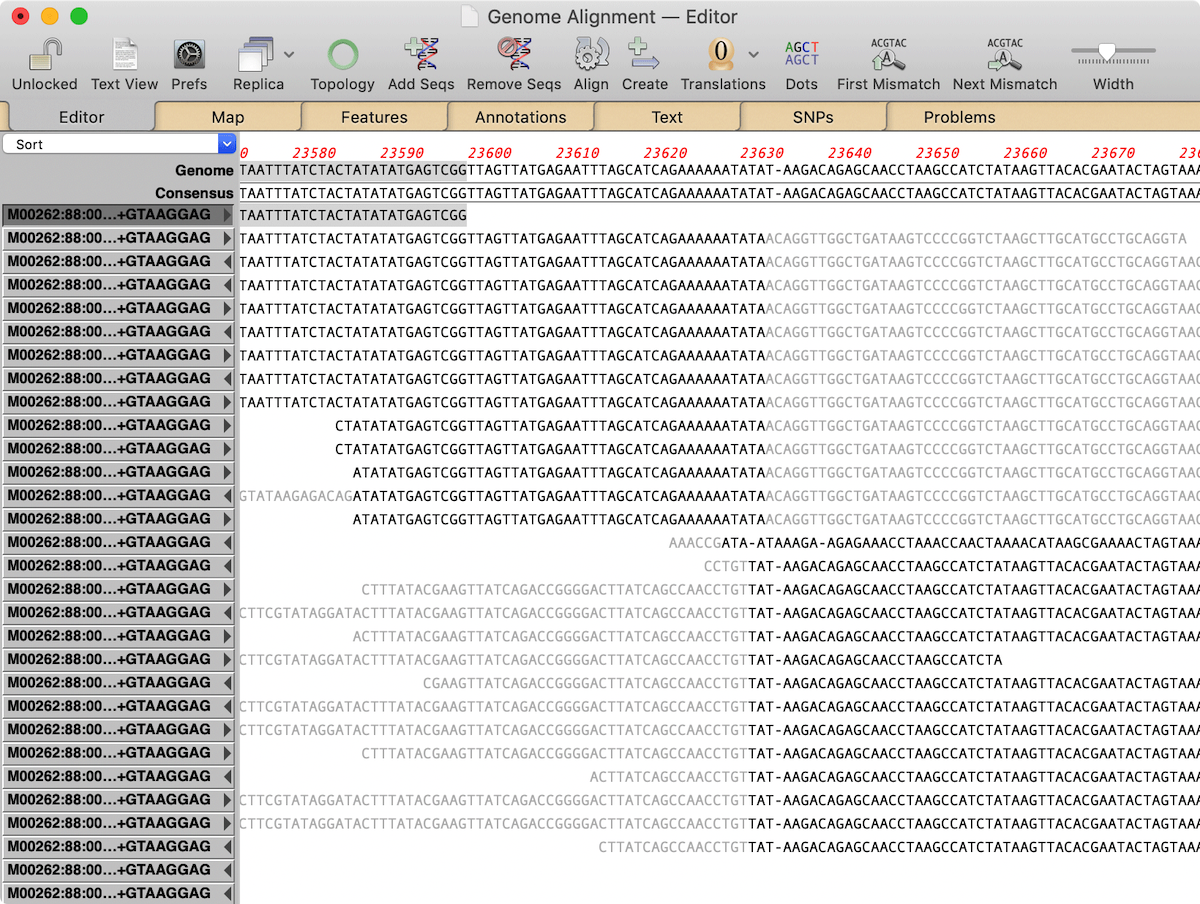
The tutorial goes into detail, describing several approaches you can use to identify the insertion locations, along with shortcuts and suggestions on how to rapidly annotate the insertion sites on the complete genome. While the tutorial does use Macvector with Assembler for parts of the analysis, you can actually accomplish the same end result using plain MacVector.