The latest releases of MacVector, MacVector 18.0.1 (Intel) and MacVector 18.1.1 (Intel and Apple Silicon) have some tweaks to the output of the SNPs tab in the Align to Reference assembly window. The genotypes of any SNP changes now follow a consistent standard, and short deletions are also reported. If the region containing the nucleotide change(s) is within an annotated CDS feature, then the genotype of the amino acid change is also reported. Here’s an example of some of the common SARS-CoV–2 variant “S” genes aligned against the reference Wuhan-HU–1 genome;
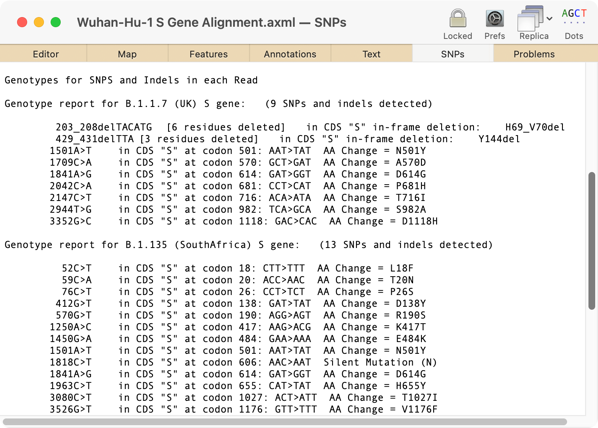
Here you can clearly see the two characteristic deletions in the B.1.1.7 UK variant – H69_V70del and Y144del along with the two amino acid changes that are believed to be critical for the increased transmission rate – N501Y and D614G. The B.1.135 South Africa variant also contains the N501Y and D614G changes, but does not have the two deletions.
MacVector 18.1 was released at the end of February and is an Universal Binary which runs on both Intel and Apple Silicon Macs. Since it follows so closely after the release of MacVector 18.0 it is not yet available through the online updater. However, if you are running an M1 Mac then you can download the installer.