With each new release of MacVector we have always strived to be the easiest to use sequence analysis application. We have made some fairly radical changes to the interface over the last few years, and inevitably there are some changes that you know will confuse long time users but are needed to make the interface more consistent and logical.
One of the major changes was in MacVector 10. Prior to this release single sequences were displayed in the sequence editor with static views of the graphical map and feature table available from this single window. The actual sequence was only represented in the editor. In MacVector 10 this changed to a tabbed window with multiple dynamic views representing the same sequence model underneath. With this more flexible approach regardless of which view you are looking at, or modifying, you are acting on the sequence directly. Furthermore because of this single sequence model all windows are linked, and you can view multiple windows (we call them replicas) of that same sequence. For example with three replicas open, one showing the sequence, another the feature table and the third the map view, then if you select a gene feature in the Feature table, then the sequence representing that gene will be highlighted, and the feature in the Map window will also be highlighted. This is great for visualising your sequences. Especially with large ones.
However, this major change has left quite a few legacy functions that do not fit the new model so well. One such function is the linear/circular button in the floating graphics palette. This button has always been a visual function, and it does not affect the real topology of the sequence underneath. There are times when it is nice to display a circular sequence in linear mode – when you want to zoom in on a crowded region for example, or looking at the MCS of a plasmid whilst cloning fragments.
This made complete sense with the old sequence model of MacVector. However with the new unified tabbed windows viewing a circular view of a linear sequence gives the incorrect impression that you are viewing a circular sequence. Thus meaning that some algorithms would provide unexpected results. A classic example is the cloning vector pBR322, which has an EcoRI site that crosses the origin. As a plasmid the sequence’s topology should be circular. However, if you have toggled the topology button in the sequence window, the sequence is now linear and the EcoR I site disappears. Prior to MacVector 11 if you now viewed this as a circular plasmid the restriction site is still absent, yet the molecule appears to be circular. That’s going to produce a confusing set of fragments on a gel!
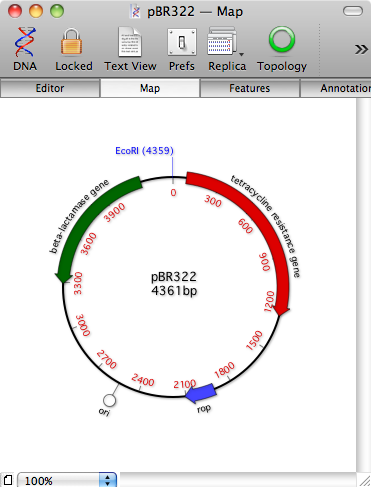
We thought that it was safer to change this behaviour in MacVector 11, and so now although you can view a circular molecule as linear, you cannot view a linear molecule as a circular one. We could not think of any user need for viewing a linear sequence as circular, although there are many advantages for viewing a circular sequence as linear. e.g. the aforementioned example of viewing the MCS of a plasmid. So now whenever you view a linear sequence the floating graphics palette linear/circular button is disabled to prevent you from inadvertently changing the Map and *thinking* you are dealing with a circular molecule. It’s a fairly small change, but unfortunately it has confused quite a few users! We do think that it is safer to have made this change. Displaying misleading results is something that we definitely do not want to do!
Currently, only the restriction enzyme, subsequence searching and (most) nucleic acid toolbox plots consider linearity/circularity in their algorithms. However, we plan on adding much better support for circular sequences in MacVector 11.5, so we thought it important to clean up this ambiguous linear/circular issue in this release. To this end, you will also see that the Topology button has now been added to the default toolbar button set of all the sequence window tabs to help indicate it is an important property of the sequence.