CRISPR-Cas9 genetic editing mechanisms require a short (typically 20nt) RNA sequence complementary to a target site next to a Protospacer Adjacent Motif (PAM) sequence. The most commonly used Cas9 nuclease (SpCas9) from Streptococcus pyogenes recognizes the PAM sequence NGG. One new function introduced a few years ago will search for PAM sequences in your sequence. There is a simple setup dialog that lets you control basic search characteristics, including choosing the specific type of nuclease to be used (we currently include 32 different characterized motifs in the default PAM file).
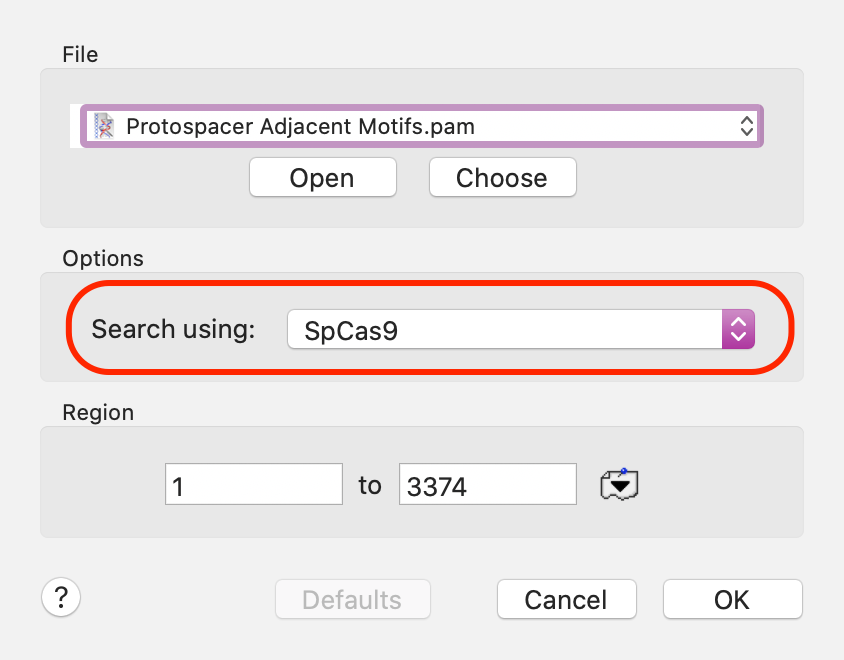
You can also search using all of the motifs in the source file, or just those selected. Potential target sites are shown graphically – here zoomed in to display the actual crRNA (lower case) and PAM sequence (upper case) of the potential target sites.

A tabular output lets you view more details about the hits.
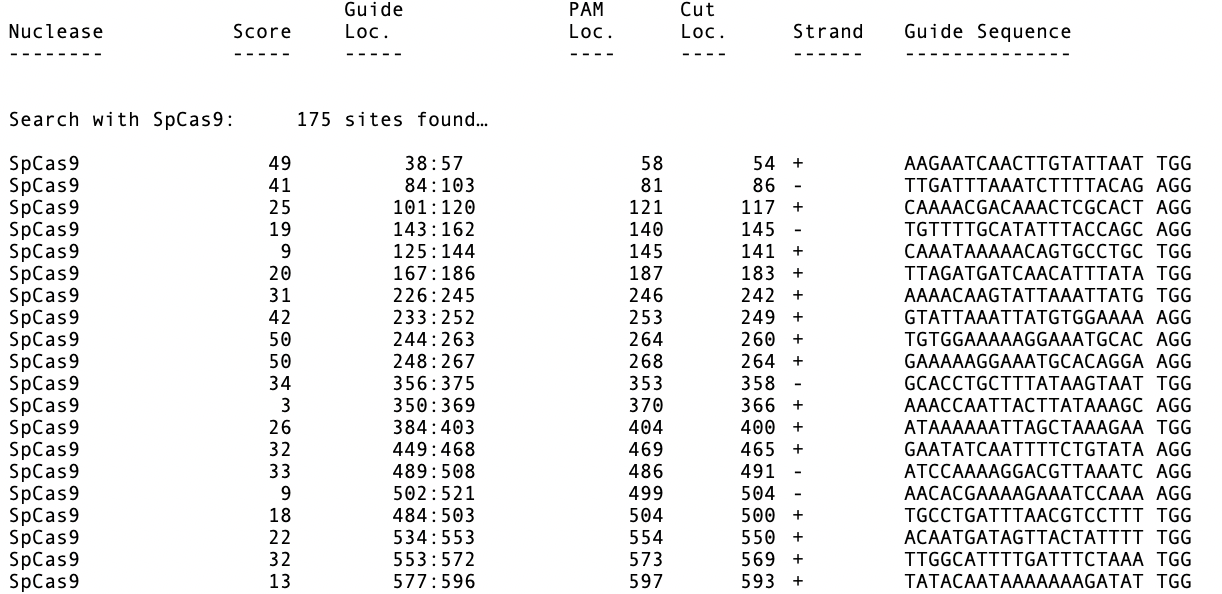
You can also configure MacVector to automatically scan all opened nucleic acid sequence documents for PAM sites using the MacVector | Preferences -> Scan DNA preference pane.