We had a recent support call this week from somebody who believed from their agarose gels that they had a mixed population of plasmids from an experiment and wanted to document and determine the banding pattern using MacVector’s agarose gel simulation. You might come across this type of scenario if you have been making site-specific mutations and introducing new restriction sites into a vector where only some of the resulting plasmids might have acquired the extra site. You can simulate this in MacVector by creating concatenated fake plasmids counting the two variant plasmids.
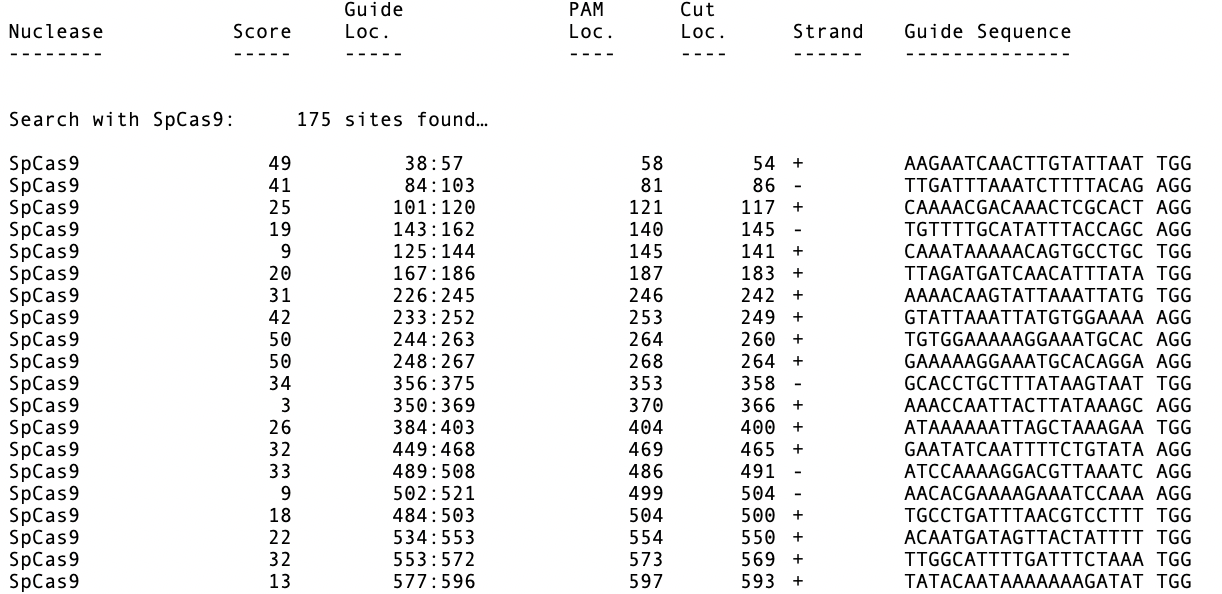
We used pBR322 and introduced a fifth FspI site by changing the T at 2788 to a C, creating pBR322+. We then joined a copy of both plasmids by selecting the unique EcoRI site in pBR322, selecting Edit | Copy, then selecting the EcoRI site in pBR322+ and choosing Edit | Paste. We saved this molecule with the two directly repeated plasmids “Hybrid–1+1”. Then we added a second copy of pBR322 into one of the EcoRI sites to create a molecule with two copies of pBR322 and one copy of pBR322+ (Hybrid–2+1). Finally we used File | New Agarose Gel to create an empty gel, then selected FspI sites from each of the four molecules and dragged them onto separate lanes in the agarose gel. (Note that by default MacVector only shows a maximum of 8 cuts per enzyme so we increased this to unlimited in the RE Picker to ensure the FspI sites were displayed).
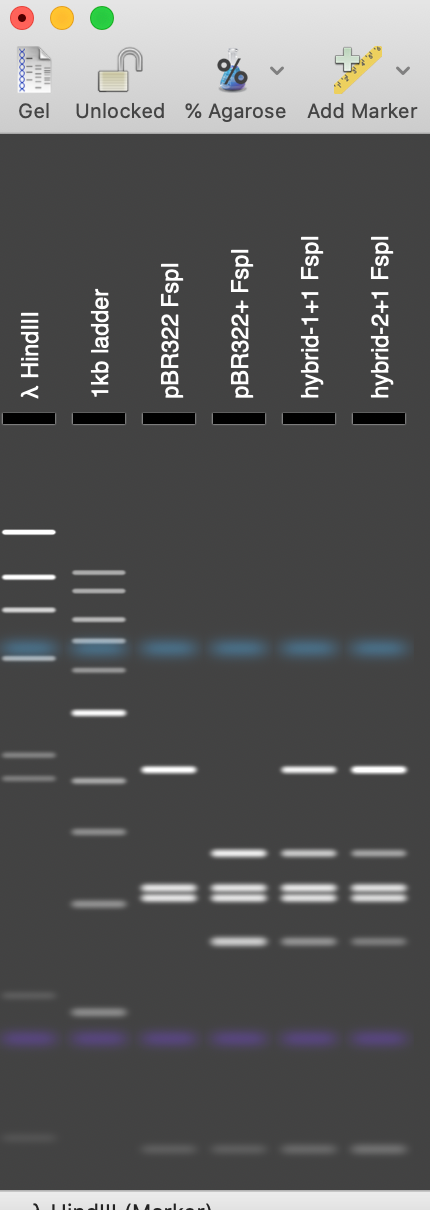
In gel tracks 3 and 4 you can see the different banding patterns between pBR322 and pBR322+. Track 5 shows the banding pattern for a 50:50 mix of the two plasmids (not the non equip-molar intensity of the variant bands) and track 6 shows the difference in band intensity when the mutant plasmid represents just one third of the molecules.